DNA losses are essential for the development of an organism. A familiar example of programmed DNA loss is the V-D-J joining in immunoglobulin genes in vertebrates that helps increase the repertoire of antibodies in mammals. In the ascarid nematodes, 'chromatin dimunition' is a form of programmed DNA loss that occurs during development at the embryonic 4 to 16 cell stage, eliminating large amounts of genomic DNA, but paradoxically adding to the total number of chromosomes. The phenomenon occurs only in five pre-somatic cells.
Although the phenomenon has been known since the 1880s, study of the fine details have only been possible in recent years after the advent of next generation sequencing. In a paper titled "Silencing of Germline-expressed genes by DNA elimination in Somatic Cells" published in the journal Developmental Cell (in 2012) , Wang et al. describe the phenomenon in Ascaris using a deep sequencing approach. The key take-aways from the paper are:
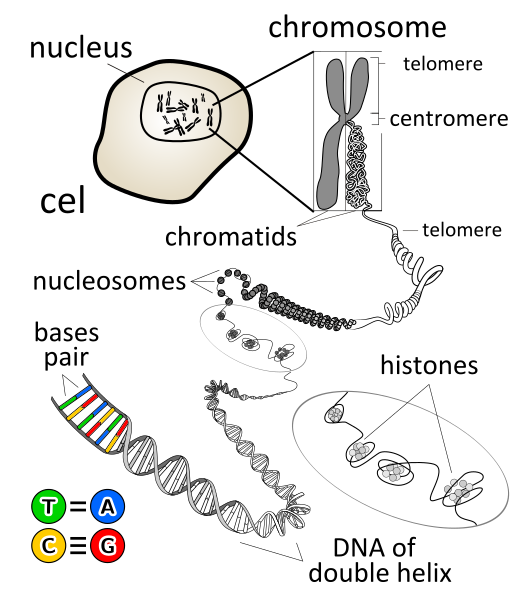
a. The sequence of events in chromatin diminution involve a break in the chromosome followed by loss of DNA and healing of the breaks by telomere addition to create new chromosomes.
b. In individual worms, the chromosomal break sites and DNA loss is pretty conserved even though there is a variation in the exact site of the break which occurs within 500 base pairs of each other in various tissues. Between individuals, this variation increases, with 70% of the breaks occurring with 1000 base pairs.
c. About 43 million base pairs are lost in total, with 12.7 million base pairs accounting for 685 unique genes expressed only during embryogenesis and/or gametogenesis.
d. Most of the eliminated genes play a role in mRNA translation and protein synthesis, and these jobs are taken up by similar "orthologous" genes in the larval and adult stages.
e. Chromatin diminution is not mediated by small RNAs like it is in ciliates, neither are there specific DNA elements in the chromosome that stipulate the break sites.
In a more recent paper titled "Comparative genome analysis of programmed DNA elimination in nematodes" published in the journal Genome Research (in 2017), Wang et al. divulge the secrets of chromatic diminution in nematodes closely related to Ascaris, namely Parascaris univalens and Toxocara canis. They show the following:
a. DNA elimination in Parascaris leads to a loss of 2.2 Gb (2.2 x 10^9 base pairs of DNA, of which 10 Mb is unique.
b. In Toxocara canis, approximately 49 Mb (49x10^6 base pairs) of DNA is eliminated, of which 20 Mb is unique.
c. The major repetitive sequences that are lost are a 120 base pairs satellite repeat in Ascaris, two short repeats of a 5 base pair sequence and a 10 base pair sequence in Parascaris and a 32-bp repeat in Toxocara canis.
d. The unique sequences that are lost include 1000 genes each in Ascaris and Parascaris, and 2000 genes in Toxocara canis. The increase in predicted genes lost in Ascaris over the previous study is of note. Since these genes are mainly expressed in the embryo, it is thought that this mechanism helps silence germline genes in nematodes.
e. The study also revealed that 86% of Ascaris genes and 88% of Parascaris genes occupy similar positions on the same chromosomes, which is also known as synteny. They also note that a set of 2623 genes are conserved between Ascaris, Parascaris univalens and Toxocara canis.
f. Chromosome break regions (CBRs) are the same in all individuals and occur at the same time in all five prismatic cells. These are regions of high SNP and indel densities. The breaks create new chromosomes, and the ends are "healed" by addition of telomeric ends. The authors identified 40, 46 and 32 CBRs in Ascaris, Parascaris and Toxocara respectively.
g. The authors were able to match 28 Ascaris CBRs to 26 CBRs in Parascaris, which is explained by synteny of genes. Such similarity did not exist in Toxocara.
h. The authors also investigated DNA accessibility in CBRs. They found no evidence of epigenetic changes but speculate that the reduced compactness of nucleosomes may lead to more accessible chromatin.
i. The authors provide preliminary evidence for changes in gene expression levels that occur as a result of proximity to telomeric ends. In Ascaris, they found that while expression levels of most genes in close proximity to the new telomeres do not change, some genes are silenced.
Both these papers describe an excellent application of the deep sequencing approach to the study of parasite biology. Chromatin diminution in nematodes, as far as we know, is an extraordinarily fascinating, precisely regulated occurrence in ascarid worms only. And as is usually the case, the more we know, the more we realize just how much we don't know yet.
References: [These are both on PMC. Click on the url to read the full articles]
Wang, J., Mitreva, M., Berriman, M., Thorne, A., Magrini, V., Koutsovoulos, G., Kumar, S., Blaxter, M. L., … Davis, R. E. (2012). Silencing of germline-expressed genes by DNA elimination in somatic cells. Developmental cell, 23(5), 1072-80. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3620533/)
Wang, J., Gao, S., Mostovoy, Y., Kang, Y., Zagoskin, M., Sun, Y., Zhang, B., White, L. K., Easton, A., Nutman, T. B., Kwok, P. Y., Hu, S., Nielsen, M. K., … Davis, R. E. (2017). Comparative genome analysis of programmed DNA elimination in nematodes. Genome research, 27(12), 2001-2014. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5741062/)
Although the phenomenon has been known since the 1880s, study of the fine details have only been possible in recent years after the advent of next generation sequencing. In a paper titled "Silencing of Germline-expressed genes by DNA elimination in Somatic Cells" published in the journal Developmental Cell (in 2012) , Wang et al. describe the phenomenon in Ascaris using a deep sequencing approach. The key take-aways from the paper are:
 |
| File:Chromosome-es.svg: KES47 (talk) derivative work: KES47 [CC BY 3.0 (https://creativecommons.org/ licenses/by/3.0)], via Wikimedia Commons |
b. In individual worms, the chromosomal break sites and DNA loss is pretty conserved even though there is a variation in the exact site of the break which occurs within 500 base pairs of each other in various tissues. Between individuals, this variation increases, with 70% of the breaks occurring with 1000 base pairs.
c. About 43 million base pairs are lost in total, with 12.7 million base pairs accounting for 685 unique genes expressed only during embryogenesis and/or gametogenesis.
d. Most of the eliminated genes play a role in mRNA translation and protein synthesis, and these jobs are taken up by similar "orthologous" genes in the larval and adult stages.
e. Chromatin diminution is not mediated by small RNAs like it is in ciliates, neither are there specific DNA elements in the chromosome that stipulate the break sites.
In a more recent paper titled "Comparative genome analysis of programmed DNA elimination in nematodes" published in the journal Genome Research (in 2017), Wang et al. divulge the secrets of chromatic diminution in nematodes closely related to Ascaris, namely Parascaris univalens and Toxocara canis. They show the following:
a. DNA elimination in Parascaris leads to a loss of 2.2 Gb (2.2 x 10^9 base pairs of DNA, of which 10 Mb is unique.
b. In Toxocara canis, approximately 49 Mb (49x10^6 base pairs) of DNA is eliminated, of which 20 Mb is unique.
c. The major repetitive sequences that are lost are a 120 base pairs satellite repeat in Ascaris, two short repeats of a 5 base pair sequence and a 10 base pair sequence in Parascaris and a 32-bp repeat in Toxocara canis.
d. The unique sequences that are lost include 1000 genes each in Ascaris and Parascaris, and 2000 genes in Toxocara canis. The increase in predicted genes lost in Ascaris over the previous study is of note. Since these genes are mainly expressed in the embryo, it is thought that this mechanism helps silence germline genes in nematodes.
e. The study also revealed that 86% of Ascaris genes and 88% of Parascaris genes occupy similar positions on the same chromosomes, which is also known as synteny. They also note that a set of 2623 genes are conserved between Ascaris, Parascaris univalens and Toxocara canis.
f. Chromosome break regions (CBRs) are the same in all individuals and occur at the same time in all five prismatic cells. These are regions of high SNP and indel densities. The breaks create new chromosomes, and the ends are "healed" by addition of telomeric ends. The authors identified 40, 46 and 32 CBRs in Ascaris, Parascaris and Toxocara respectively.
g. The authors were able to match 28 Ascaris CBRs to 26 CBRs in Parascaris, which is explained by synteny of genes. Such similarity did not exist in Toxocara.
h. The authors also investigated DNA accessibility in CBRs. They found no evidence of epigenetic changes but speculate that the reduced compactness of nucleosomes may lead to more accessible chromatin.
i. The authors provide preliminary evidence for changes in gene expression levels that occur as a result of proximity to telomeric ends. In Ascaris, they found that while expression levels of most genes in close proximity to the new telomeres do not change, some genes are silenced.
Both these papers describe an excellent application of the deep sequencing approach to the study of parasite biology. Chromatin diminution in nematodes, as far as we know, is an extraordinarily fascinating, precisely regulated occurrence in ascarid worms only. And as is usually the case, the more we know, the more we realize just how much we don't know yet.
References: [These are both on PMC. Click on the url to read the full articles]
Wang, J., Mitreva, M., Berriman, M., Thorne, A., Magrini, V., Koutsovoulos, G., Kumar, S., Blaxter, M. L., … Davis, R. E. (2012). Silencing of germline-expressed genes by DNA elimination in somatic cells. Developmental cell, 23(5), 1072-80. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3620533/)
Wang, J., Gao, S., Mostovoy, Y., Kang, Y., Zagoskin, M., Sun, Y., Zhang, B., White, L. K., Easton, A., Nutman, T. B., Kwok, P. Y., Hu, S., Nielsen, M. K., … Davis, R. E. (2017). Comparative genome analysis of programmed DNA elimination in nematodes. Genome research, 27(12), 2001-2014. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5741062/)